Selected Projects
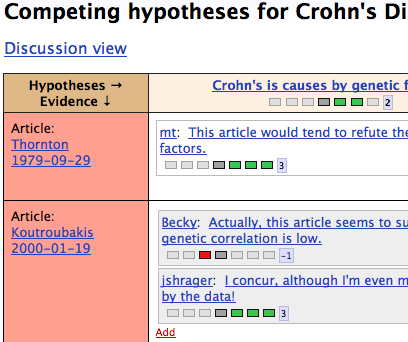
BRUCE
data portal for brain cancer researchData portal for BRUCE project - BRain tUmor heterogeneity deCiphEred by high dimensional multiomic analysis.
Enflame
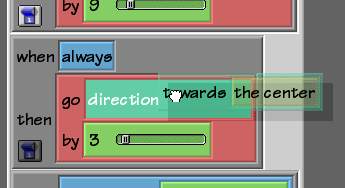
visual query builderA Scratch-like visual query builder for scientific graph knowledge bases. Core components released as the open-source Blockoid library.
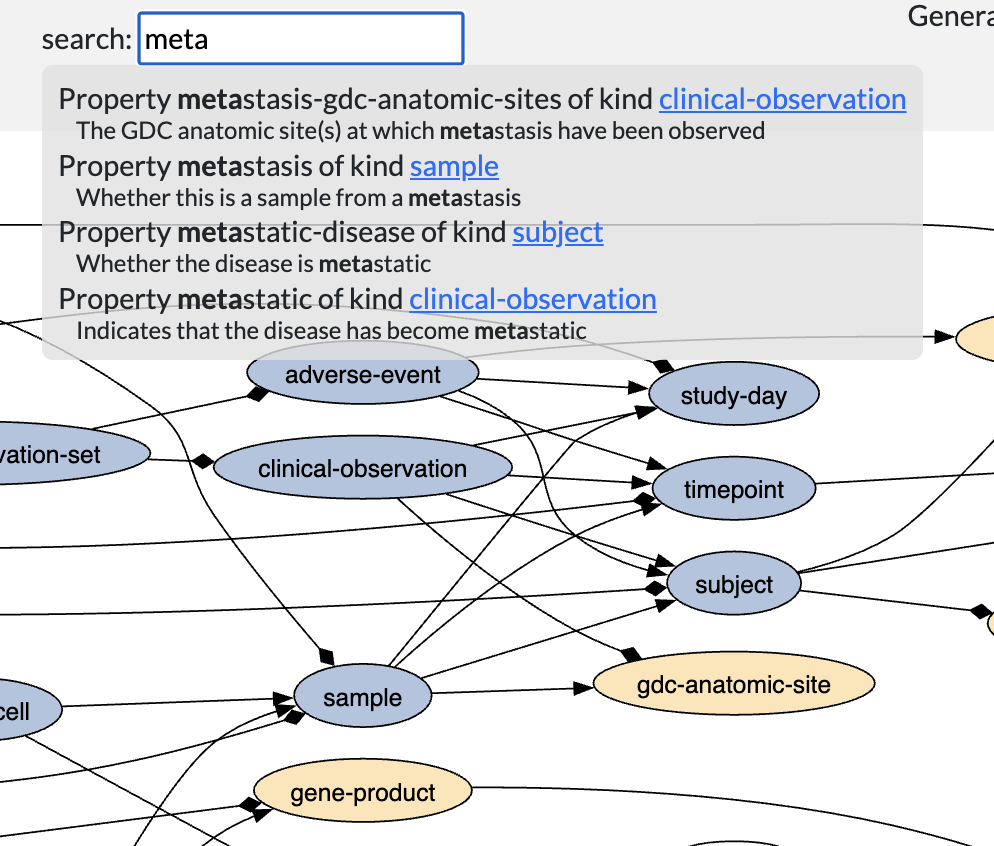
Alzabo
schema management toolOntology management tool and documentation generator for graph databases
SmartTables
semantic spreadsheetA semantic spreadsheet system for bioinformatics, deployed in production at BioCyc for biological research data management.
Goddinpotty
digital garden frameworkA tool to generate static sites (personal wikis or digital gardens) from Logseq or other graph-based knowledge managers.